H0: σπ2 = 0
H1: σπ2 <> 0
Assumptions
| 1. | Independence |
| 2. | Normality |
| 3. | Homogeneity of variance |
| 4. | No Non-additivity (no block*treatment interaction) |
| 5. | Compound symmetry in the variance-covariance matrix |
ANOVA Summary Table
| Source | SS | df | MS | F | p-value | Error | |
| 1 | Treatment | 49.00 | 3 | 16.33 | 11.63 | 0.0001 | [3] |
| 2 | Blocks (Subjects) | 31.50 | 7 | 4.50 | 3.20 | 0.0180 | [3] |
| 3 | Residual | 29.50 | 21 | 1.40 | |||
| Total | 110.00 | 31 |
Table of the F-distribution
Expected Mean Squares
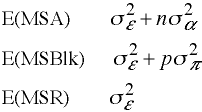
Strength of Association
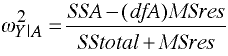
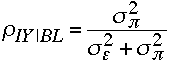
Using Stata: Method 1
input y a s x1 x2 x3 s1 s2 s3 s4 s5 s6 s7
3 1 1 1 1 1 1 1 1 1 1 1 1
2 1 2 1 1 1 -1 1 1 1 1 1 1
2 1 3 1 1 1 0 -2 1 1 1 1 1
3 1 4 1 1 1 0 0 -3 1 1 1 1
1 1 5 1 1 1 0 0 0 -4 1 1 1
3 1 6 1 1 1 0 0 0 0 -5 1 1
4 1 7 1 1 1 0 0 0 0 0 -6 1
6 1 8 1 1 1 0 0 0 0 0 0 -7
4 2 1 -1 1 1 1 1 1 1 1 1 1
4 2 2 -1 1 1 -1 1 1 1 1 1 1
3 2 3 -1 1 1 0 -2 1 1 1 1 1
3 2 4 -1 1 1 0 0 -3 1 1 1 1
2 2 5 -1 1 1 0 0 0 -4 1 1 1
3 2 6 -1 1 1 0 0 0 0 -5 1 1
4 2 7 -1 1 1 0 0 0 0 0 -6 1
5 2 8 -1 1 1 0 0 0 0 0 0 -7
4 3 1 0 -2 1 1 1 1 1 1 1 1
4 3 2 0 -2 1 -1 1 1 1 1 1 1
3 3 3 0 -2 1 0 -2 1 1 1 1 1
3 3 4 0 -2 1 0 0 -3 1 1 1 1
4 3 5 0 -2 1 0 0 0 -4 1 1 1
6 3 6 0 -2 1 0 0 0 0 -5 1 1
5 3 7 0 -2 1 0 0 0 0 0 -6 1
5 3 8 0 -2 1 0 0 0 0 0 0 -7
3 4 1 0 0 -3 1 1 1 1 1 1 1
5 4 2 0 0 -3 -1 1 1 1 1 1 1
6 4 3 0 0 -3 0 -2 1 1 1 1 1
5 4 4 0 0 -3 0 0 -3 1 1 1 1
7 4 5 0 0 -3 0 0 0 -4 1 1 1
6 4 6 0 0 -3 0 0 0 0 -5 1 1
10 4 7 0 0 -3 0 0 0 0 0 -6 1
8 4 8 0 0 -3 0 0 0 0 0 0 -7
end
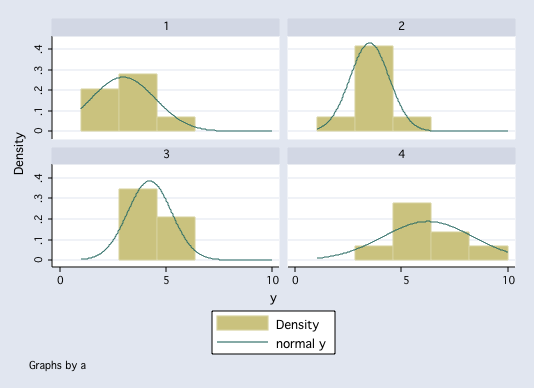
tabstat y, by(a) stat(n mean sd var)
Summary for variables: y
by categories of: a
a | N mean sd variance
---------+----------------------------------------
1 | 8 3 1.511858 2.285714
2 | 8 3.5 .9258201 .8571429
3 | 8 4.25 1.035098 1.071429
4 | 8 6.25 2.12132 4.5
---------+----------------------------------------
Total | 32 4.25 1.883716 3.548387
--------------------------------------------------
histogram y, by(a) normal
 anova y a s, repeated(a)
Number of obs = 32 R-squared = 0.7318
Root MSE = 1.18523 Adj R-squared = 0.6041
Source | Partial SS df MS F Prob > F
-----------+----------------------------------------------------
Model | 80.50 10 8.05 5.73 0.0004
|
a | 49.00 3 16.3333333 11.63 0.0001
s | 31.50 7 4.50 3.20 0.0180
|
Residual | 29.50 21 1.4047619
-----------+----------------------------------------------------
Total | 110.00 31 3.5483871
Between-subjects error term: s
Levels: 8 (7 df)
Lowest b.s.e. variable: s
Repeated variable: a
Huynh-Feldt epsilon = 0.8343
Greenhouse-Geisser epsilon = 0.6195
Box's conservative epsilon = 0.3333
------------ Prob > F ------------
Source | df F Regular H-F G-G Box
-----------+----------------------------------------------------
a | 3 11.63 0.0001 0.0003 0.0015 0.0113
Residual | 21
-----------+----------------------------------------------------
mat list e(Srep)
symmetric e(Srep)[4,4]
c1 c2 c3 c4
r1 2.2857143
r2 1.1428571 .85714286
r3 .71428571 .28571429 1.0714286
r4 1.2857143 .28571429 .92857143 4.5
effectsize a
anova effect size for a with dep var = y
total variance accounted for
omega2 = .40200898
eta2 = .44545455
Cohen's f = .81991823
partial variance accounted for
partial omega2 = .49907137
partial eta2 = .62420382
nonadd y a s
Tukey's test of nonadditivity for randomized block designs
Ho: model is additive (no block*treatment interaction)
F (1,20) = 1.2795813 Pr > F: .27135918
quietly anova y a s /* quietly rerun anova to get correct mse */
tkcomp a
Tukey-Kramer pairwise comparisons for variable a
studentized range critical value(.05, 4, 21) = 3.9419483
mean
grp vs grp group means dif TK-test
-------------------------------------------------------
1 vs 2 3.0000 3.5000 0.5000 1.1932
1 vs 3 3.0000 4.2500 1.2500 2.9830
1 vs 4 3.0000 6.2500 3.2500 7.7558*
2 vs 3 3.5000 4.2500 0.7500 1.7898
2 vs 4 3.5000 6.2500 2.7500 6.5626*
3 vs 4 4.2500 6.2500 2.0000 4.7728*
tukeyhsd a
Tukey HSD pairwise comparisons for variable a
studentized range critical value(.05, 4, 21) = 3.9419483
uses harmonica mean sample size = 8.000
mean critical
grp vs grp group means dif dif
-------------------------------------------------------
1 vs 2 3.0000 3.5000 0.5000 1.6518
1 vs 3 3.0000 4.2500 1.2500 1.6518
1 vs 4 3.0000 6.2500 3.2500* 1.6518
2 vs 3 3.5000 4.2500 0.7500 1.6518
2 vs 4 3.5000 6.2500 2.7500* 1.6518
3 vs 4 4.2500 6.2500 2.0000* 1.6518
regress y x1 x2 x3 s1 s2 s3 s4 s5 s6 s7
Source | SS df MS Number of obs = 32
-------------+------------------------------ F( 10, 21) = 5.73
Model | 80.50 10 8.05 Prob > F = 0.0004
Residual | 29.50 21 1.4047619 R-squared = 0.7318
-------------+------------------------------ Adj R-squared = 0.6041
Total | 110.00 31 3.5483871 Root MSE = 1.1852
------------------------------------------------------------------------------
y | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
x1 | -.25 .2963066 -0.84 0.408 -.8662034 .3662034
x2 | -.3333333 .1710727 -1.95 0.065 -.6890985 .0224318
x3 | -.6666667 .1209667 -5.51 0.000 -.9182306 -.4151027
s1 | -.125 .4190409 -0.30 0.768 -.9964432 .7464432
s2 | .0416667 .2419334 0.17 0.865 -.4614613 .5447946
s3 | .0208333 .1710727 0.12 0.904 -.3349318 .3765985
s4 | .0125 .1325124 0.09 0.926 -.2630745 .2880745
s5 | -.1583333 .1081959 -1.46 0.158 -.383339 .0666723
s6 | -.2916667 .0914422 -3.19 0.004 -.4818312 -.1015022
s7 | -.25 .0791913 -3.16 0.005 -.4146873 -.0853127
_cons | 4.25 .2095204 20.28 0.000 3.814278 4.685722
------------------------------------------------------------------------------
test x1 x2 x3
( 1) x1 = 0.0
( 2) x2 = 0.0
( 3) x3 = 0.0
F( 3, 21) = 11.63
Prob > F = 0.0001
test s1 s2 s3 s4 s5 s6 s7
( 1) s1 = 0.0
( 2) s2 = 0.0
( 3) s3 = 0.0
( 4) s4 = 0.0
( 5) s5 = 0.0
( 6) s6 = 0.0
( 7) s7 = 0.0
F( 7, 21) = 3.20
Prob > F = 0.0180
regress y i.a i.s
Source | SS df MS Number of obs = 32
-------------+------------------------------ F( 10, 21) = 5.73
Model | 80.5 10 8.05 Prob > F = 0.0004
Residual | 29.5 21 1.4047619 R-squared = 0.7318
-------------+------------------------------ Adj R-squared = 0.6041
Total | 110 31 3.5483871 Root MSE = 1.1852
------------------------------------------------------------------------------
y | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
a |
2 | .5 .5926133 0.84 0.408 -.7324067 1.732407
3 | 1.25 .5926133 2.11 0.047 .0175933 2.482407
4 | 3.25 .5926133 5.48 0.000 2.017593 4.482407
|
s |
2 | .25 .8380817 0.30 0.768 -1.492886 1.992886
3 | -1.78e-16 .8380817 -0.00 1.000 -1.742886 1.742886
4 | -3.41e-16 .8380817 -0.00 1.000 -1.742886 1.742886
5 | -1.86e-16 .8380817 -0.00 1.000 -1.742886 1.742886
6 | 1 .8380817 1.19 0.246 -.7428863 2.742886
7 | 2.25 .8380817 2.68 0.014 .5071137 3.992886
8 | 2.5 .8380817 2.98 0.007 .7571137 4.242886
|
_cons | 2.25 .6949006 3.24 0.004 .804875 3.695125
------------------------------------------------------------------------------
/* user written program -- findit anovalator */
anovalator a s, main fratio
anovalator main-effect for a
chi2(3) = 34.881356 p-value = 1.291e-07
scaled as F-ratio = 11.627119
anovalator main-effect for s
chi2(7) = 22.423729 p-value = .00214632
scaled as F-ratio = 3.2033898
anova y a s, repeated(a)
Number of obs = 32 R-squared = 0.7318
Root MSE = 1.18523 Adj R-squared = 0.6041
Source | Partial SS df MS F Prob > F
-----------+----------------------------------------------------
Model | 80.50 10 8.05 5.73 0.0004
|
a | 49.00 3 16.3333333 11.63 0.0001
s | 31.50 7 4.50 3.20 0.0180
|
Residual | 29.50 21 1.4047619
-----------+----------------------------------------------------
Total | 110.00 31 3.5483871
Between-subjects error term: s
Levels: 8 (7 df)
Lowest b.s.e. variable: s
Repeated variable: a
Huynh-Feldt epsilon = 0.8343
Greenhouse-Geisser epsilon = 0.6195
Box's conservative epsilon = 0.3333
------------ Prob > F ------------
Source | df F Regular H-F G-G Box
-----------+----------------------------------------------------
a | 3 11.63 0.0001 0.0003 0.0015 0.0113
Residual | 21
-----------+----------------------------------------------------
mat list e(Srep)
symmetric e(Srep)[4,4]
c1 c2 c3 c4
r1 2.2857143
r2 1.1428571 .85714286
r3 .71428571 .28571429 1.0714286
r4 1.2857143 .28571429 .92857143 4.5
effectsize a
anova effect size for a with dep var = y
total variance accounted for
omega2 = .40200898
eta2 = .44545455
Cohen's f = .81991823
partial variance accounted for
partial omega2 = .49907137
partial eta2 = .62420382
nonadd y a s
Tukey's test of nonadditivity for randomized block designs
Ho: model is additive (no block*treatment interaction)
F (1,20) = 1.2795813 Pr > F: .27135918
quietly anova y a s /* quietly rerun anova to get correct mse */
tkcomp a
Tukey-Kramer pairwise comparisons for variable a
studentized range critical value(.05, 4, 21) = 3.9419483
mean
grp vs grp group means dif TK-test
-------------------------------------------------------
1 vs 2 3.0000 3.5000 0.5000 1.1932
1 vs 3 3.0000 4.2500 1.2500 2.9830
1 vs 4 3.0000 6.2500 3.2500 7.7558*
2 vs 3 3.5000 4.2500 0.7500 1.7898
2 vs 4 3.5000 6.2500 2.7500 6.5626*
3 vs 4 4.2500 6.2500 2.0000 4.7728*
tukeyhsd a
Tukey HSD pairwise comparisons for variable a
studentized range critical value(.05, 4, 21) = 3.9419483
uses harmonica mean sample size = 8.000
mean critical
grp vs grp group means dif dif
-------------------------------------------------------
1 vs 2 3.0000 3.5000 0.5000 1.6518
1 vs 3 3.0000 4.2500 1.2500 1.6518
1 vs 4 3.0000 6.2500 3.2500* 1.6518
2 vs 3 3.5000 4.2500 0.7500 1.6518
2 vs 4 3.5000 6.2500 2.7500* 1.6518
3 vs 4 4.2500 6.2500 2.0000* 1.6518
regress y x1 x2 x3 s1 s2 s3 s4 s5 s6 s7
Source | SS df MS Number of obs = 32
-------------+------------------------------ F( 10, 21) = 5.73
Model | 80.50 10 8.05 Prob > F = 0.0004
Residual | 29.50 21 1.4047619 R-squared = 0.7318
-------------+------------------------------ Adj R-squared = 0.6041
Total | 110.00 31 3.5483871 Root MSE = 1.1852
------------------------------------------------------------------------------
y | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
x1 | -.25 .2963066 -0.84 0.408 -.8662034 .3662034
x2 | -.3333333 .1710727 -1.95 0.065 -.6890985 .0224318
x3 | -.6666667 .1209667 -5.51 0.000 -.9182306 -.4151027
s1 | -.125 .4190409 -0.30 0.768 -.9964432 .7464432
s2 | .0416667 .2419334 0.17 0.865 -.4614613 .5447946
s3 | .0208333 .1710727 0.12 0.904 -.3349318 .3765985
s4 | .0125 .1325124 0.09 0.926 -.2630745 .2880745
s5 | -.1583333 .1081959 -1.46 0.158 -.383339 .0666723
s6 | -.2916667 .0914422 -3.19 0.004 -.4818312 -.1015022
s7 | -.25 .0791913 -3.16 0.005 -.4146873 -.0853127
_cons | 4.25 .2095204 20.28 0.000 3.814278 4.685722
------------------------------------------------------------------------------
test x1 x2 x3
( 1) x1 = 0.0
( 2) x2 = 0.0
( 3) x3 = 0.0
F( 3, 21) = 11.63
Prob > F = 0.0001
test s1 s2 s3 s4 s5 s6 s7
( 1) s1 = 0.0
( 2) s2 = 0.0
( 3) s3 = 0.0
( 4) s4 = 0.0
( 5) s5 = 0.0
( 6) s6 = 0.0
( 7) s7 = 0.0
F( 7, 21) = 3.20
Prob > F = 0.0180
regress y i.a i.s
Source | SS df MS Number of obs = 32
-------------+------------------------------ F( 10, 21) = 5.73
Model | 80.5 10 8.05 Prob > F = 0.0004
Residual | 29.5 21 1.4047619 R-squared = 0.7318
-------------+------------------------------ Adj R-squared = 0.6041
Total | 110 31 3.5483871 Root MSE = 1.1852
------------------------------------------------------------------------------
y | Coef. Std. Err. t P>|t| [95% Conf. Interval]
-------------+----------------------------------------------------------------
a |
2 | .5 .5926133 0.84 0.408 -.7324067 1.732407
3 | 1.25 .5926133 2.11 0.047 .0175933 2.482407
4 | 3.25 .5926133 5.48 0.000 2.017593 4.482407
|
s |
2 | .25 .8380817 0.30 0.768 -1.492886 1.992886
3 | -1.78e-16 .8380817 -0.00 1.000 -1.742886 1.742886
4 | -3.41e-16 .8380817 -0.00 1.000 -1.742886 1.742886
5 | -1.86e-16 .8380817 -0.00 1.000 -1.742886 1.742886
6 | 1 .8380817 1.19 0.246 -.7428863 2.742886
7 | 2.25 .8380817 2.68 0.014 .5071137 3.992886
8 | 2.5 .8380817 2.98 0.007 .7571137 4.242886
|
_cons | 2.25 .6949006 3.24 0.004 .804875 3.695125
------------------------------------------------------------------------------
/* user written program -- findit anovalator */
anovalator a s, main fratio
anovalator main-effect for a
chi2(3) = 34.881356 p-value = 1.291e-07
scaled as F-ratio = 11.627119
anovalator main-effect for s
chi2(7) = 22.423729 p-value = .00214632
scaled as F-ratio = 3.2033898
Using Stata: Method 2
Randomized block data often come in a wide form, in which, each of the repeated measures is a separate variable. Before you can alanlyze these data using the Stata anova command you need to reshape the data into a long form. While the data are still wide you can compute the correlation and covariance matrices using the corr command.
input s y1 y2 y3 y4
1 3 4 4 3
2 2 4 4 5
3 2 3 3 6
4 3 3 3 5
5 1 2 4 7
6 3 3 6 6
7 4 4 5 10
8 6 5 5 8
end
corr y1 y2 y3 y4, cov /* note: covariance matrix same as e(Srep) */
(obs=8)
| y1 y2 y3 y4
---------+------------------------------------
y1 | 2.28571
y2 | 1.14286 .857143
y3 | .714286 .285714 1.07143
y4 | 1.28571 .285714 .928571 4.5
corr y1 y2 y3 y4
(obs=8)
| y1 y2 y3 y4
-------------+------------------------------------
y1 | 1.0000
y2 | 0.8165 1.0000
y3 | 0.4564 0.2981 1.0000
y4 | 0.4009 0.1455 0.4229 1.0000
reshape long y, i(s) j(a)
(note: j = 1 2 3 4)
Data wide -> long
-----------------------------------------------------------------------------
Number of obs. 8 -> 32
Number of variables 5 -> 3
j variable (4 values) -> a
xij variables:
y1 y2 ... y4 -> y
-----------------------------------------------------------------------------
list
[output omitted]
anova y a s, repeated(a)
Number of obs = 32 R-squared = 0.7318
Root MSE = 1.18523 Adj R-squared = 0.6041
Source | Partial SS df MS F Prob > F
-----------+----------------------------------------------------
Model | 80.50 10 8.05 5.73 0.0004
|
a | 49.00 3 16.3333333 11.63 0.0001
s | 31.50 7 4.50 3.20 0.0180
|
Residual | 29.50 21 1.4047619
-----------+----------------------------------------------------
Total | 110.00 31 3.5483871
Between-subjects error term: s
Levels: 8 (7 df)
Lowest b.s.e. variable: s
Repeated variable: a
Huynh-Feldt epsilon = 0.8343
Greenhouse-Geisser epsilon = 0.6195
Box's conservative epsilon = 0.3333
------------ Prob > F ------------
Source | df F Regular H-F G-G Box
-----------+----------------------------------------------------
a | 3 11.63 0.0001 0.0003 0.0015 0.0113
Residual | 21
-----------+----------------------------------------------------
Nonadditivity